pymatgen 使用
pymatgen 使用
介绍
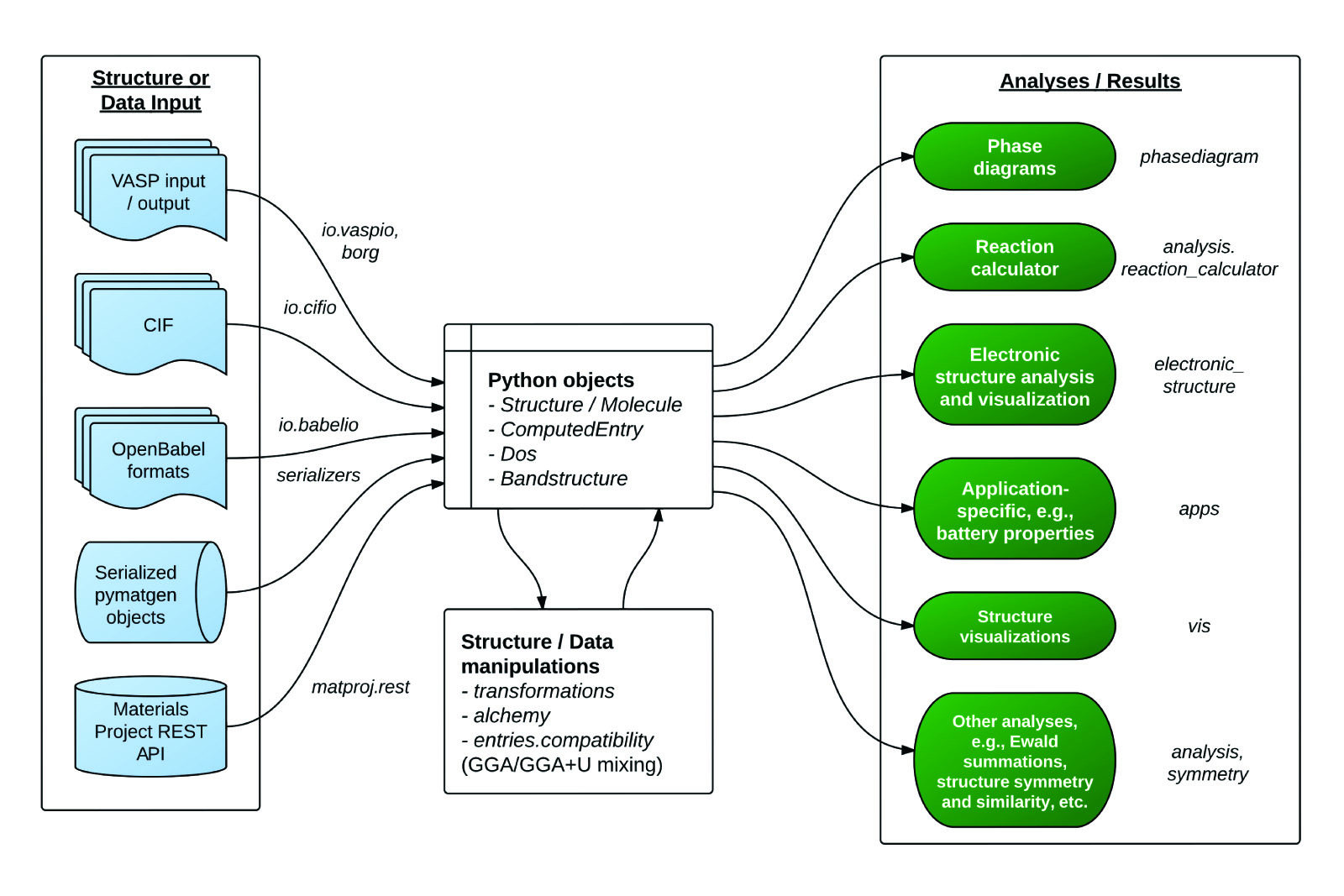
用于表示 Element、Site、Structure、Molecule 的高度灵活的类
文件输入/输出支持广泛,如 VASP、ABINIT、CIF、Gaussian、XYZ 等(主要依靠 Open Babel 包)
强大的分析工具,包括生成相图、Pourbaix 图、扩散分析、反应等
电子结构分析,如态密度和能带结构
集成 Materials Project REST API、Crystallography Open Database 等其他外部数据源
代码文档详细
参考资料
pymatgen notebook:GitHub - yw-fang/pymatgen-notebook
pymatgen 实例代码:GitHub - materialsvirtuallab/matgenb
Materials Project 官方文档:Materials Project Documentation
- Materials Project 推荐赝势:Pseudo-potentials - Materials Project Documentation
- MP 计算所采用的具体参数及其说明(如截断能为 520eV 是由元素周期表所有元素赝势中最大截断能的 1.3 倍得到的): Materials Methodology - Materials Project Documentation
GitHub - computron/pymatgen_tutorials: Tutorials for using the pymatgen library
Material Project Workshop:
- 2021:The Materials Project Workshop
- 2018~2020:Releases · materialsproject/workshop
- 2017:GitHub - materialsproject/workshop-2017: Assets for the 2017 Materials Project workshop
- 2016:GitHub - materialsproject/workshop-2016: Assets for the Materials Project workshop in Aug 2016
- 注:Workshop 2020 和 2021 的内容绝大部分相似,lesson3 分别为表面和界面;Workshop 2018 和 2019 的内容相似(对 atomate 的讲解稍微详细些)
安装
安装
1 | # 稳定版本 |
赝势目录设置
change log(代码 bug 修复,新功能添加等,可以关注)
兼容性:需对进行
from pymatgen import xxx修改(v2022.0.0 版本开始)
1 | # 旧 |
使用
基本
查看 pymatgen 相关信息
1 | import pymatgen.core |
- 获取 .pmgrc 配置文件内容
1 | from pymatgen.core import SETTINGS |
- pymatgen 支持的构型文件格式
1 | # pymatgen/core/structure.py |
- pymatgen 中可指定的泛函类型
1 | # pymatgen/io/vasp/sets.py |
MP 晶体 DFT code 用的是 VASP,分子用的是 Q-Chem
MSONable类:MSON(Monty JSON);MSONable 对象必须实现as_dict()方法,该方法须返回可序列化为 JSON 的字典,且须支持无参数;以及实现from_dict()类方法,即从as_dict()方法生成的字典中重建对象;as_dict()方法应该包含@module和@class键,这将允许 MontyEncoder 动态反序列化该类monty 包:对 json/yaml/msgpack 等文件格式进行序列化(比 json 模块好用很多,推荐!)
1 | from monty.serialization import loadfn, dumpfn |
工作流
- pymatgen 典型工作流
CLI
- 不是太好用,建议直接写脚本
1 | # 查看子命令帮助 |
- pymatgen 的 cli 可使用 argcomplete 库(
pyamtgen/cli/pmg.py;用于 cli 命令的补全)
1 | try: |
操作 structure
- 包括创建、保存、分析与变化操作
1 | from pymatgen.core.composition import Composition |
Entry
- Entry 的最基本形式:一个构型成分 + 对应计算的能量(可包含其他输入或计算数据);可作为 pymatgen 创建相图的计算数据集
计算输入输出管理
pymatgen.io 模块包含一些计算软件(主要是 VASP)的输入文件编写与解析和输出文件解析子模块
输入管理的核心类是 InputSet(如 VaspInputSet)。InputSet 对象包含计算输入文件所需的所有数据。可使用
write_input()方法将所有输入文件生成到指定路径;可使用from_directory()静态方法从计算目录中构建 InputSet许多解析输出文件的类继承自 InputFile,其提供了一个读写文件的标准接口
其他
1 | # 可视化原子构型 |
- 生成元素置换后的非等同结构
- 借助 bsym 包;适用于复杂结构,如金属间化合物;可理解为置换找到的非等同原子中等同原子的第一个原子
- bsym_examples
1 | from bsym.interface.pymatgen import unique_structure_substitutions |
- 解析 VASP 计算目录:Automated DFT - The Materials Project Workshop
1 | from atomate.vasp.drones import VaspDrone |
- 静电势能计算
- 【Pymatgen学习 2】Ewald方法计算静电能
- Ewald Summation - Qijing Zheng
- EwaldSummation 是 pymatgen 库中的一个类,用于计算离子晶体的 Ewald 总能量。Ewald 总能量是一种用于处理带电体系的长程库仑相互作用的技术,通常用于计算固体材料中的电势能。该方法将总能量分解为实空间、倒空间、点电荷修正和偶极修正部分,并进行相应的求和计算
求助:过渡态计算新版pymatgen中找不到iddp插值方法 - 第一性原理 (First Principle) - 计算化学公社
1 | # 过渡态 NEB |
相关问题
pymatgen 如何获取可用的 POTCAR 种类;较难:一般是指定泛函类型
pymatgen structure 如何通过 structure 来生成 potcar?解决方法:通过 Poscar 类得到 structure 的元素种类,之后与 PBE 泛函的元素进行比对,之后用 Potcar 类写入 POTCAR(生成新的之前需删掉原来的 POTCAR 文件)
pymatgen 中的 LLL reduction 是什么含义
pymatgen 键长计算(并非只是简单的计算原子对之间的距离)
pymatgen 保存成 POSCAR 时,对元素类型进行排序(是否可自定义);不方便,建议用 ase ✅ 2024-10-23
-
to()或Poscar类中的write_file()方法无直接排序参数 ✅ 2024-10-23 -
Structure类中的sort()方法中的key参数可以是哪些值(不深挖) ✅ 2024-10-23
-
常用模块
pymatgen.core
- pymatgen 核心模块,常用子模块:structure、sites、 lattice、 composition、 periodic_table
structure
晶体结构模块
Structure 类继承自 IStructure
Structure 与 Molecule 类的区别
- Molecule 类的基本参数为
species、coords,Structure 类还需指定lattice参数 - Molecule 类的
coords参数值需是笛卡尔坐标形式,Structure 类可以是笛卡尔和分数两种坐标形式 - Molecule 类本质上是 Site objects 的列表;Structure 类本质上是 PeriodicSites objects 的列表;两者均可类似 list 一样进行操作
- Molecule 类的基本参数为
查看方法是否会直接修改对象本身:检查是否有
in_place参数及注释是否有 “in place” 字样(“原地” 的意思)Structure 类无
wrap()方法(根据 PBC 将胞外原子移至胞内),ASE 有:pymatgen - What Does the coordinate list next to the cartesian coordinates of an atom represent in neighbor_list - Stack Overflow复杂结构无法通过
to_primitive()方法(以及 vaspkit)将单胞转化成原胞Structure 类相关属性和方法:
1 | from pymatgen.core.structure import Structure |
composition
- 成分/化学式模块
1 | from pymatgen.core.composition import Composition |
periodic_table
- 元素周期表模块,查看元素信息/数据
1 | from pymatgen.core.periodic_table import Element |
sites
- 原子位点模块
1 | from pymatgen.core.sites import Site, PeriodicSite |
lattice
- 点阵模块
1 | from pymatgen.core.lattice import Lattice |
units
- pymatgen 相关工具
1 | from pymatgen.util.string import ... |
bonds
- 键
1 | from pymatgen.core.bonds import get_bond_length |
surface
表面 slab 模块;总体而言,该模块生成表面模型没有 ASE 相关模块或 atomsk 或 latgen 好用
1 | from pymatge.core.surface import ... |
SlabGenerator类相关属性和方法:
1 | from pymatge.core.surface import SlabGenerator |
pymatgen.io.ase
ASE 接口
AseAtomsAdaptor:将 ASE 中的Atoms类与 pymatgen 中的Structure类互相转换
1 | from pymatgen.io.ase import AseAtomsAdaptor |
pymatgen.io.atat
ATAT 接口
只有 Mcsqs 类(功能较一般)
pymatgen.io.phonopy
- Phonopy 接口
1 | from pymatgen.io.phonopy import get_phonopy_structure, get_pmg_structure |
pymatgen.io.vasp.help
- 查看 VASP 参数 help
1 | from pymatgen.io.vasp.help import VaspDoc |
pymatgen.io.vasp.inputs
VASP 输入文件模块
四种类
Incar、Poscar、Kpoints、Potcar;都有from_dict()、from_file()、write_file()类方法
Incar
- 在解析 INCAR 文件时,得到的字典的键和值都是字符串,需要对 INCAR 中不同参数的键的值的类型进行正确的转换,因此定义了
proc_val()函数
1 | from pymatgen.io.vasp.inputs import Incar |
Kpoints
生成 K 点密度的 classmethod 大多都有
force_gamma参数automatic_density_by_vol()类方法生成的三个方向的 K 点密度公式可理解为: reciprocal_density * (2*π / lattice_constant);kppvol参数值设置可参考MPStaticSet类中的代码
1 | from pymatgen.io.vasp.inputs import Kpoints |
Poscar
1 | from pymatgen.io.vasp.inputs import Poscar |
Potcar
- 读取和写入 POTCAR 文件的 object,由
PotcarSingleobject (单个 POTCAR) 的列表组成
1 | from pymatgen.io.vasp.inputs import Potcar |
pymatgen.io.vasp.sets
VaspInputSet类
1 | from pymatgen.io.vasp.sets import VaspInputSet |
- 不同 project 的 VASP 输入文件参数设置 yaml 文件
1 | # 弛豫计算;MP 默认的输入文件参数设置 |
MPRelaxSet、MPStaticSet等类均继承于VaspInputSet(很重要,多仔细阅读其 docstring)MPRelaxSet没有设置 EDIFFG 参数:MPRelaxSet.write_input() no EDIFFG in INCAR - pymatgen - Materials Science Community DiscourseMPRelaxSet中的赝势选择相比 MIT project 参数,含更多的价电子;K 点网格密度也比其高 50%在 MPRelaxSet.yaml 参数设置文件中,KPOINTS 只能写如下几种参数
1 | # 在 VaspInputSet 类的 kpoints 属性中查看 |
- 常见计算类型 Set
1 | # MP 表示 Matetials Project |
- MPRelaxSet.yaml 文件主要参数设置
1 | PARENT: VASPIncarBase |
- VaspInputSet 类中对 INCAR LMAXMIX 参数的设置(VASP 默认值为 2)
1 | # pymatgen/io/vasp/sets.py 第 644 ~ 656 行 |
MVLNPTMDSetNPT 系综 AIMD 参数设置
1 | # pymatgen/io/vasp/sets.py |
pymatgen.io.vasp.outputs
- 读取并解析 VASP 的输出文件
Outcar
- Outcar 类能获取的较普适数据的属性和方法较少(主要解析 Vasprun.xml 文件无法获取到的数据)
1 | from pymatgen.io.vasp.ouputs import Outcar |
Oszicar
- Oszicar 类的
final_energy属性选择的是E0;Vasprun 类的final_energy属性选择的也是E0
1 | from pymatgen.io.vasp.ouputs import Oszicar |
Vasprun
BSVasprun 类:Vasprun 的优化版本(继承至 Vasprun),只解析能带结构的本征值(忽略结构,参数等)
1 | from pymatgen.io.vasp.ouputs import Vasprun |
- Vasprun 类在解析未收敛的计算时,会有 Warning,并说明电子步和离子步的收敛情况
1 | UnconvergedVASPWarning: vasprun.xml is an unconverged VASP run. |
Vaspout
- 解析 vaspout.h5 文件(pymatgen 2025.03.10 版本支持);需安装 h5py 包
1 | from pymatgen.io.vasp.outputs import Vaspout |
其他
1 | from pymatgen.io.vasp.outputs import Elfcar, Chgcar |
pymatgen.io.lammps
data
- 解析 LAMMPS data 格式文件
1 | from pymatgen.io.lammps.data import LammpsData |
pymatgen.io.phonopy
1 | from pymatgen.io.phonopy import get_phonopy_structure, get_pmg_structure |
pymatgen.electronic_structure
电子结构相关工具与分析(能带和态密度);可绘制能带、DOS、能带 + DOS
Plotter 类的 sigma 参数使绘制出的图平滑(sigma=0.05 相对好一些;若平滑后使原本的部分数据信息损失,建议不设置该参数!)
态密度
态密度 DOS 数据获取与绘图
- label 颜色自定义:修改源代码(获取 ax 的方法不行)
- 使用 pymatgen 模块绘制 DOS 图时,相对会耗时一些,建议将 DOS 数据获取后单独存储为数据文件自己绘制
1 | import matplotlib.pyplot as plt |
能带
1 | import matplotlib.pyplot as plt |
COHP
- COHP (Crystal orbital Hamilton population) 与 ICOHP 之间的区别是什么
sampleScript/RHEAs/COHP.py at master · Jeffery-Li0513/sampleScript · GitHub
1 | from pymatgen.electronic_structure.cohp import Cohp, get_integrated_cohp_in_energy_range |
pymatgen.analysis
structure_matcher
- 查看两个结构之间的相似度(是否相同/接近)
1 | from pymatgen.analysis.structure_matcher import StructureMatcher |
eos
- EOS 类:
BirchMurnaghan、Birch、Murnaghan、PourierTarantola、Vinet等
1 | from pymatgen.analysis.eos import BirchMurnaghan |
interface
Working with Surfaces and Interfaces - The Materials Project Workshop
pymatgen 界面模型生成无法指定两种 slab 的层数:https://matsci.org/t/building-a-slab-and-interface/45317
生成所有可能匹配的超晶格的过程:
- 减少表面/界面(surfaces)晶格向量并计算表面/界面的面积
- 在最大允许面积内生成所有超晶格变换
- 对于每个超晶格集:
- 减少超晶格矢量
- 检查 film 和 substrate 表面超晶格之间的长度和角度
1 | from pymatgen.analysis.interfaces.coherent_interfaces import CoherentInterfaceBuilder |
elasticity
1 | from pymatgen.analysis.elasticity import ElasticTensor, Strain, Stress |
electronic_structure
- pymatgen 电子结构相关分析很多都是建立在 vasprun.xml 文件中提取数据之上的(与 vaspkit 有不同)
phase_diagram
可绘制二、三、四元的相图
无法直接使用
pymatgen.entries中的Entry类初始化,会报错,energy参数为ABC抽象类型(Doc 有提及),而是用PDEntry或ComputedEntry类初始化Entry中的 composition 参数值尽量不用约化的化学式;energy 参数值为该成分体系对应的总能量 eV,非已计算的形成能 eV/atomlabel 字体大小无法调节:How control fontsize in PDPlotter? - pymatgen - Materials Science Community Discourse
1 | # 绘制相图 Convex Hull |
diffraction
- XRD 绘制:
1 | from pymatgen.analysis.diffraction.xrd import XRDCalculator |
adsorption
1 | from pymatgen.analysis.adsorption import AdsorbateSiteFinder, plot_slab |
prototype
- 匹配晶体结构的 prototype(调用的是 AFLOW 的 prototype 数据)
1 | from pymatgen.analysis.prototype import ... |
diffusion
1 | pip install -U pymatgen-analysis-diffusion |
- 使用
1 | # 使用 IDPP 方法进行插值 |
pymatgen.symmetry.bandstructure
- 寻找高对称 K 点
1 | from pymatgen.symmetry.bandstructure import HighSymmKpath |
pymatgen.symmetry.analyzer
SpacegroupAnalyzer
- 空间群分析
1 | from pymatgen.symmetry.analyzer import SpacegroupAnalyzer |
pymatgen.transformations
参考:
简单的变换操作:添加和删除原子位点,元素替换,到更高级的一对多的转换
pymatgen 无置换 Wyckoff Site 中的元素的类
SQSTransformation类中调用的是 ATAT 中的 mcsqs 工具或 ICET 中的 enumeration、monte carlo 模块;建议直接使用其调用的原生工具
1 | from pymatgen.transformations.standard_transformations import ... |
pymatgen.phonon
1 | from pymatgen.phonon.bandstructure import PhononBandStructureSymmLine |